Allelica’s ancestry first approach for clinical PRS implementation
George Busby, Allelica CSO & Co-Founder
Polygenic Risk Scores (PRS) are a powerful tool in precision medicine, offering insights into an individual’s genetic predisposition to various diseases. The accuracy and applicability of PRSs can be significantly influenced by genetic ancestry, posing challenges in their use across diverse populations. This article explores the intricacies of PRS, the impact of genetic ancestry, and the methods we use to overcome these challenges.
Recap on Polygenic Risk Scores
First, a quick reminder that Polygenic Risk Scores (PRSs) are calculated by aggregating the effects of numerous genetic variants across the genome. These scores estimate an individual’s genetic risk for common diseases such as breast cancer, type 2 diabetes, and coronary artery disease. The utility of PRS lies in its potential to inform personalized healthcare strategies, enabling early intervention and tailored prevention measures.
Despite their promise, a valid concern is that because PRS models predominantly rely on data from genome-wide association studies (GWAS), which are heavily skewed towards individuals of European ancestry, the resulting PRSs can be less accurate when applied to individuals from other ancestral backgrounds. The lack of transferability of PRSs developed on European populations into other groups has been noted by many authors and has necessitated a more inclusive approach to genetic research, which aims to increase the representation of non-European ancestry individuals in clinical datasets.
Why PRSs developed in one group often do not transfer to other populations
Recall that an individual’s PRS value is calculated by assessing their genotypes at a set of variants in a PRS panel. The panel comprises a list of variants, each of which has a risk allele and associated effect size. The PRS is calculated as a sum of all the variant-level effects present in an individual, with these variant level effects computed based on the number of risk alleles they carry at each variant.

A key assumption, which we always validate, is that the risk allele and associated effect on disease represent true effects on disease.
There are several population genetic reasons why this assumption might not hold across different populations, which can be summarised as follows:
- Differences in allele frequencies in different populations mean that some risk alleles are more common in some populations. This can lead to inflated PRS values not because there is inherently more risk in these populations, but because risk alleles are more common.
- Some variants in PRS panels tag functional changes but are not causal of disease themselves. Because patterns of Linkage Disequilibrium vary in different populations, a variant that is associated with risk in one population, might not be as closely associated with risk in another.
- The magnitude of the effect on disease of an allele in one population might differ in different populations. Even if a risk allele accurately captures risk in different populations, its effect on disease can vary.
All three of these explanations can be related to differences in the genetic architecture of disease, which is captured by assessments of genetic ancestry across populations. We recently wrote about genetic ancestry, and how it can more accurately be described as genetic similarity.
It therefore follows that if we can account for and correct differences in genetic ancestry, then we can go a long way to ameliorate the observed attenuation of PRS performance across different populations.
Addressing this challenge requires innovative approaches to model development and data collection.
Methods to Account for Genetic Ancestry Differences
Several methods have been developed to mitigate genetic ancestry bias in PRSs, all of which we deploy at Allelica to ensure that our clinical PRSs are available for all:
Ancestry-Specific PRS Models
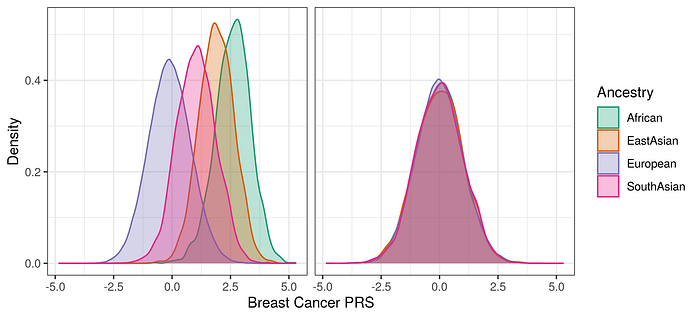
Ancestry-specific models tailor PRS to specific populations by using GWAS and PRS validation data from those groups. As we showed in our most recent coronary artery disease and breast cancer PRSs, (Busby et al 2023; Busby et al under review), this approach improves accuracy by aligning the genetic variants considered in the PRS with the target population’s genetic architecture. However, this approach requires diverse GWAS datasets, the dearth of which remains a significant hurdle.
Genetic Ancestry Adjustment Techniques
Raw PRS values can be adjusted using genetic ancestry inferred through principal components. These methods enhance the performance of PRS by removing the influence of allele frequency differences between populations.
Optimising PRS panels to include functional variants
Finemapping and other techniques aim to focus genetic effects onto variants that are more likely to be causal of disease, rather than being variants that simply tag the causal variant. The overall effect of this approach is to use more putatively causal variants in a PRS panel, which reduces (or even removes) the attenuation in performance that is caused by differences in LD patterns between different populations.
Utilizing genetic ancestry specific populations for PRS testing and validation
To provide accurate assessments of PRS in different populations, it is essential to test their performance in different populations. This explicitly acknowledges that PRS performance varies in different groups, but aims to establish a more accurate translation of PRS into risk reports by assessing the performance of the PRS in different groups. Allelica use ancestry specific risk distributions to accurately predict risk.
Ancestry-specific approaches improve clinical PRSs
These advancements in PRS development mean that even compared to a few years ago, we can be more confident that our clinical PRSs can be accurately applied to the populations that we serve. The underrepresentation of non-European populations in genetic research is still an issue, and we support all efforts to increase representation. The continued development of diverse biobanks (e.g. All of Us) will greatly aid these endeavours. Expanding the diversity of genetic datasets is crucial for equitable healthcare.
You can read much more detail about these developments in our recent papers (Busby et al 2023; Busby et al under review). The application of polygenic risk scores across different genetic ancestries raises complexities due to population biases, genetic diversity, and differing environmental influences. However, by diversifying genomic data, employing multiancestry models, and utilizing advanced machine learning and functional genomics, Allelica has developed accurate and universally applicable PRS. These efforts ensure that the benefits of genetic research and personalized medicine are equitably distributed across all populations.
